Note
Click here to download the full example code
Model 3 - Recumbent Fold¶
A recumbent (overturned) fold. We start by importing the necessary dependencies:
Importing GemPy
import gempy as gp
# Importing auxiliary libraries
import pandas as pd
pd.set_option('precision', 2)
Creating the model by importing the input data and displaying it:
data_path = 'https://raw.githubusercontent.com/cgre-aachen/gempy_data/master/'
path_to_data = data_path + "/data/input_data/jan_models/"
geo_data = gp.create_data('recumbent',
extent=[0, 1000, 0, 1000, 0, 1000], resolution=[50, 50, 50],
path_o=path_to_data + "model3_orientations.csv",
path_i=path_to_data + "model3_surface_points.csv")
Out:
Active grids: ['regular']
Setting and ordering the units and series:
gp.map_stack_to_surfaces(geo_data, {"Strat_Series": ('rock2', 'rock1'), "Basement_Series": ('basement')})
gp.plot_2d(geo_data, direction=['y'])
Out:
<gempy.plot.visualization_2d.Plot2D object at 0x7fcc6bbb8b80>
Calculating the model:
interp_data = gp.set_interpolator(geo_data, theano_optimizer='fast_compile')
Out:
Setting kriging parameters to their default values.
Compiling theano function...
Level of Optimization: fast_compile
Device: cpu
Precision: float64
Number of faults: 0
Compilation Done!
Kriging values:
values
range 1732.05
$C_o$ 71428.57
drift equations [3, 3]
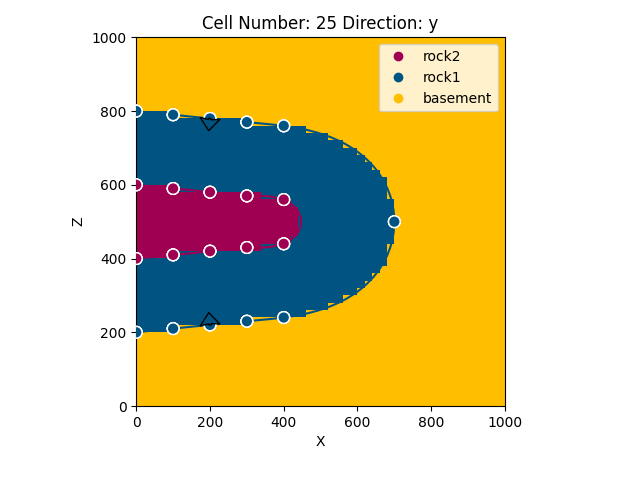
Displaying the result in x and y direction:
sphinx_gallery_thumbnail_number = 2
gp.plot_2d(geo_data, cell_number=[25],
direction=['y'], show_data=True)
gp.save_model(geo_data)
Out:
True
Total running time of the script: ( 0 minutes 9.014 seconds)
